Accessing versioned data¶
The Delphi Epidata API stores not just each signal’s estimate for a given location on a given day, but also when that estimate was made, and all updates to that estimate.
For example, let’s look at the doctor visits signal from the covidcast endpoint, which estimates the percentage of outpatient doctor visits that are COVID-related.
Consider a result row with time_value = 2020-05-01 for geo_values = "pa". This is an estimate for Pennsylvania on May 1, 2020. That estimate was issued on May 5, 2020 (which is recorded in the issue column), the delay coming from a combination of:
time taken by our data partner to collect the data
time taken by the Dekohu Epidata API to ingest the data provided.
Later, the estimate for May 1st could be updated, perhaps because additional visit data from May 1st arrived at our source and was reported to us. This constitutes a new issue of the data.
Data known “as of” a specific date¶
By default, endpoint functions fetch the most recent issue available. This is the best option for users who simply want to graph the latest data or construct dashboards. But if we are interested in knowing when data was reported, we can request specific data versions using the as_of, issues, or lag arguments (note that these are mutually exclusive and that not all endpoints aside from pub_covidcast support all three parameters, so please check the documentation for that
specific endpoint).
First, we can request the data that was available as it was available on a specific date, using the as_of argument:
from epidatpy import EpiDataContext, EpiRange
epidata = EpiDataContext(use_cache=False)
# Obtain the most up-to-date version of the smoothed covid-like illness (CLI)
# signal from the COVID-19 Trends and Impact survey for the US
epidata.pub_covidcast(
data_source="doctor-visits",
signals="smoothed_cli",
time_type="day",
time_values="2020-05-01",
geo_type="state",
geo_values="pa",
as_of="2020-05-07",
).df()
| source | signal | geo_type | geo_value | time_type | time_value | issue | lag | value | stderr | sample_size | direction | missing_value | missing_stderr | missing_sample_size | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | doctor-visits | smoothed_cli | state | pa | day | 2020-05-01 | 2020-05-07 | 6 | 2.32192 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
This shows that an estimate of about 2.3% was issued on May 7. If we don’t specify as_of, we get the most recent estimate available:
epidata.pub_covidcast(
data_source="doctor-visits",
signals="smoothed_cli",
time_type="day",
time_values="2020-05-01",
geo_type="state",
geo_values="pa",
).df()
| source | signal | geo_type | geo_value | time_type | time_value | issue | lag | value | stderr | sample_size | direction | missing_value | missing_stderr | missing_sample_size | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | doctor-visits | smoothed_cli | state | pa | day | 2020-05-01 | 2020-07-04 | 64 | 5.075015 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
Note the substantial change in the estimate, from less than 3% to over 5%, reflecting new data that became available after May 7 about visits occurring on May 1. This illustrates the importance of issue date tracking, particularly for forecasting tasks. To backtest a forecasting model on past data, it is important to use the data that would have been available at the time the model was or would have been fit, not data that arrived much later.
By plotting API results with different values of the as_of parameter, we can see how the indicator value changes over time as new observations become available:
import matplotlib.pyplot as plt
plt.rcParams["figure.dpi"] = 300
results = []
for as_of_date in ["2020-05-07", "2020-05-14", "2020-05-21", "2020-05-28"]:
apicall = epidata.pub_covidcast(
data_source="doctor-visits",
signals="smoothed_adj_cli",
time_type="day",
time_values=EpiRange("2020-04-20", "2020-04-27"),
geo_type="state",
geo_values="pa",
as_of=as_of_date,
)
results.append(apicall.df())
final_df = pd.concat(results)
final_df["issue"] = final_df["issue"].dt.date
fig, ax = plt.subplots(figsize=(6, 5))
ax.spines["right"].set_visible(False)
ax.spines["left"].set_visible(False)
ax.spines["top"].set_visible(False)
final_df.pivot_table(values="value", index="time_value", columns="issue").plot(
xlabel="Date", ylabel="CLI", ax=ax, linewidth=1.5
)
plt.title("Smoothed CLI from Doctor Visits", fontsize=16)
plt.subplots_adjust(bottom=0.2)
plt.show()
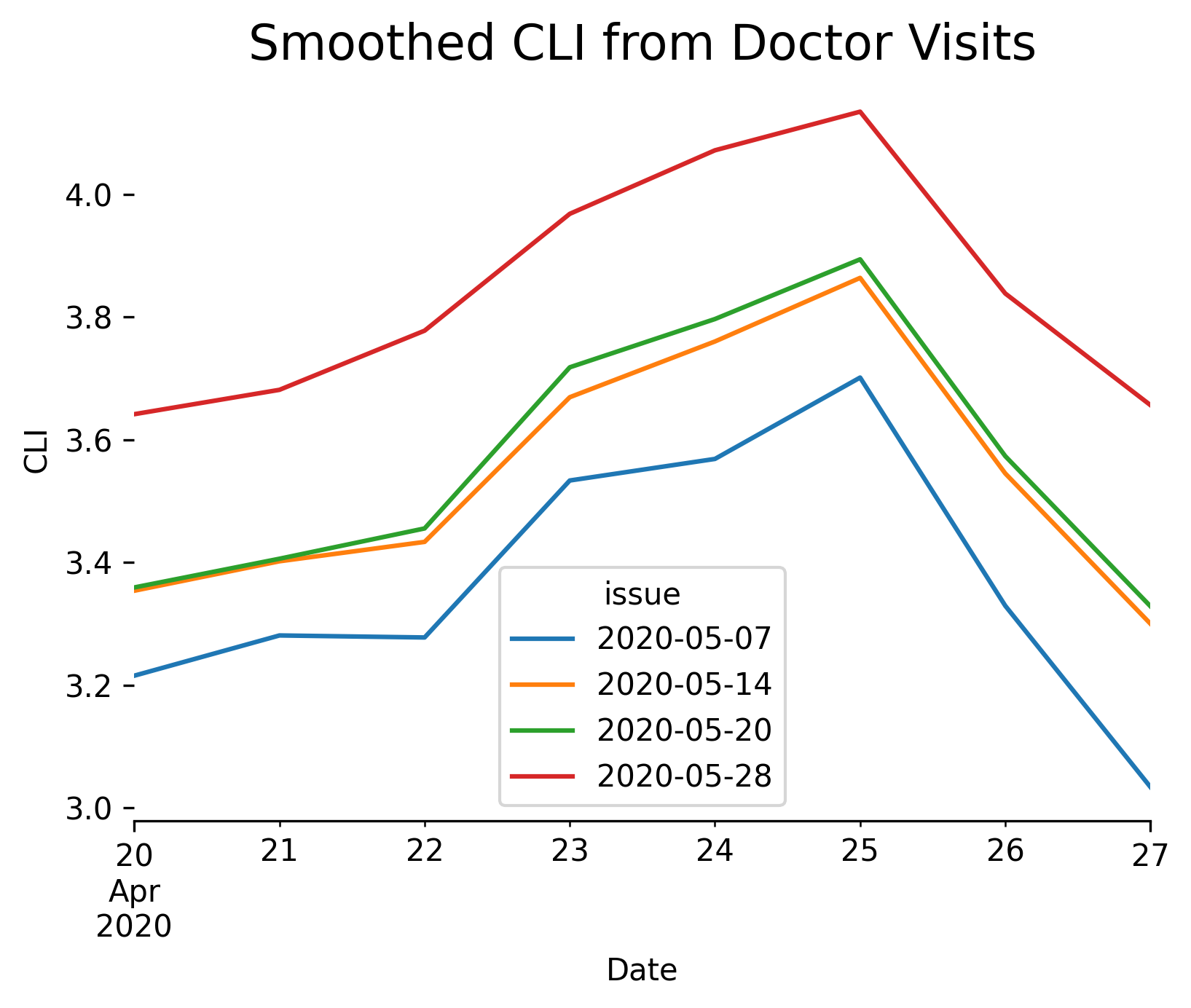
Multiple issues of observations¶
By using the issues argument, we can request all issues in a certain time period:
epidata.pub_covidcast(
data_source="doctor-visits",
signals="smoothed_adj_cli",
time_type="day",
time_values="2020-05-01",
geo_type="state",
geo_values="pa",
issues=EpiRange("2020-05-01", "2020-05-15"),
).df()
| source | signal | geo_type | geo_value | time_type | time_value | issue | lag | value | stderr | sample_size | direction | missing_value | missing_stderr | missing_sample_size | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | doctor-visits | smoothed_adj_cli | state | pa | day | 2020-05-01 | 2020-05-07 | 6 | 2.581509 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
| 1 | doctor-visits | smoothed_adj_cli | state | pa | day | 2020-05-01 | 2020-05-08 | 7 | 3.278896 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
| 2 | doctor-visits | smoothed_adj_cli | state | pa | day | 2020-05-01 | 2020-05-09 | 8 | 3.321781 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
| 3 | doctor-visits | smoothed_adj_cli | state | pa | day | 2020-05-01 | 2020-05-12 | 11 | 3.588683 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
| 4 | doctor-visits | smoothed_adj_cli | state | pa | day | 2020-05-01 | 2020-05-13 | 12 | 3.631978 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
| 5 | doctor-visits | smoothed_adj_cli | state | pa | day | 2020-05-01 | 2020-05-14 | 13 | 3.658009 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
| 6 | doctor-visits | smoothed_adj_cli | state | pa | day | 2020-05-01 | 2020-05-15 | 14 | 3.662286 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
This estimate was clearly updated many times as new data for May 1st arrived. Note that these results include only data issued or updated between (inclusive) 2020-05-01 and 2020-05-15. If a value was first reported on 2020-04-15, and never updated, a query for issues between 2020-05-01 and 2020-05-15 will not include that value among its results. This view of the data is useful for understanding the revision patterns in a signal and can be useful for nowcasting (i.e. the practice of auto-correcting real-time estimates).
Observations issued with a specific lag¶
Finally, we can use the lag argument to request only data reported with a certain lag. For example, requesting a lag of 7 days fetches only data issued exactly 7 days after the corresponding time_value:
epidata.pub_covidcast(
data_source="doctor-visits",
signals="smoothed_adj_cli",
time_type="day",
time_values=EpiRange("2020-05-01", "2020-05-01"),
geo_type="state",
geo_values="pa",
lag=7,
).df()
| source | signal | geo_type | geo_value | time_type | time_value | issue | lag | value | stderr | sample_size | direction | missing_value | missing_stderr | missing_sample_size | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | doctor-visits | smoothed_adj_cli | state | pa | day | 2020-05-01 | 2020-05-08 | 7 | 3.278896 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
Note that though this query requested all values between 2020-05-01 and 2020-05-07, May 3rd and May 4th were not included in the results set. This is because the query will only include a result for May 3rd if a value were issued on May 10th (a 7-day lag), but in fact the value was not updated on that day:
epidata.pub_covidcast(
data_source="doctor-visits",
signals="smoothed_adj_cli",
time_type="day",
time_values="2020-05-03",
geo_type="state",
geo_values="pa",
issues=EpiRange("2020-05-09", "2020-05-15"),
).df()
| source | signal | geo_type | geo_value | time_type | time_value | issue | lag | value | stderr | sample_size | direction | missing_value | missing_stderr | missing_sample_size | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | doctor-visits | smoothed_adj_cli | state | pa | day | 2020-05-03 | 2020-05-09 | 6 | 2.788618 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
| 1 | doctor-visits | smoothed_adj_cli | state | pa | day | 2020-05-03 | 2020-05-12 | 9 | 3.015368 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
| 2 | doctor-visits | smoothed_adj_cli | state | pa | day | 2020-05-03 | 2020-05-13 | 10 | 3.03931 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
| 3 | doctor-visits | smoothed_adj_cli | state | pa | day | 2020-05-03 | 2020-05-14 | 11 | 3.021245 | <NA> | <NA> | <NA> | 0 | 5 | 5 |
| 4 | doctor-visits | smoothed_adj_cli | state | pa | day | 2020-05-03 | 2020-05-15 | 12 | 3.048725 | <NA> | <NA> | <NA> | 0 | 5 | 5 |