4. Combine external data sources and COVIDcast data
Source:vignettes/external-data.Rmd
external-data.RmdWhile the COVIDcast Epidata API provides numerous useful COVID data
streams, you may sometimes find yourself with relevant data from a
different source. This package provides the tools you need to load such
data and use it alongside COVIDcast data—for example, you can calculate
correlations with covidcast_cor() and make maps using
plot.covidcast_signal().
Let’s illustrate this in action using data from the COVID Tracking Project. They provide CSV files for download containing daily state-level data on numerous metrics, including cases, deaths, hospitalizations, and test results.
Loading the data
We’ve saved a sample of the data in this package to use as an
example. Let’s load it and examine a few columns using
knitr::kable() to print them nicely:
library(dplyr)
data <- read.csv(system.file("extdata", "covid-tracking-project-oct-2020.csv",
package = "covidcast", mustWork = TRUE))
data %>%
select(date, state, death, deathIncrease, hospitalizedCurrently,
hospitalizedIncrease) %>%
head() %>%
knitr::kable()| date | state | death | deathIncrease | hospitalizedCurrently | hospitalizedIncrease |
|---|---|---|---|---|---|
| 2020-10-31 | AK | 82 | 1 | 94 | 7 |
| 2020-10-31 | AL | 2967 | 35 | 960 | 0 |
| 2020-10-31 | AR | 1925 | 25 | 652 | 35 |
| 2020-10-31 | AS | 0 | 0 | NA | 0 |
| 2020-10-31 | AZ | 5979 | 45 | 889 | 68 |
| 2020-10-31 | CA | 17626 | 55 | 3212 | 0 |
This is in a convenient format: Each day’s observations for each
state are in one row. Suppose we would like to extract
hospitalizedIncrease as a signal we want to map and analyze
alongside other data fetched with covidcast_signal().
To do this, we use as.covidcast_signal(). It expects a
data frame with at least three columns:
-
time_value: aDateobject giving the observation date -
value: the value of the observation, such as the number of deaths or hospitalizations -
geo_value: the location, such as state or county, in the same form as returned bycovidcast_signal()(such as two-letter lowercase abbreviations for states and FIPS codes for counties)
Other columns are preserved unchanged. (In particular, if an
issue column is present, it is used as the issue date of
each observation. This is important if your data source includes
multiple revisions of each observation.)
as.covidcast_signal() also needs to know the
geo_type (state, in this case) and source/signal name to
apply to the data. With a bit of dplyr data wrangling, we can do
this easily:
library(covidcast)
hospitalized <- data %>%
select(time_value = date,
geo_value = state,
value = hospitalizedIncrease) %>%
mutate(geo_value = tolower(geo_value),
time_value = as.Date(time_value)) %>%
as.covidcast_signal(geo_type = "state",
data_source = "covid-tracking",
signal = "hospitalized_increase")
head(hospitalized) %>%
knitr::kable()| data_source | signal | geo_value | time_value | value | issue |
|---|---|---|---|---|---|
| covid-tracking | hospitalized_increase | ak | 2020-10-31 | 7 | 2023-07-12 |
| covid-tracking | hospitalized_increase | al | 2020-10-31 | 0 | 2023-07-12 |
| covid-tracking | hospitalized_increase | ar | 2020-10-31 | 35 | 2023-07-12 |
| covid-tracking | hospitalized_increase | as | 2020-10-31 | 0 | 2023-07-12 |
| covid-tracking | hospitalized_increase | az | 2020-10-31 | 68 | 2023-07-12 |
| covid-tracking | hospitalized_increase | ca | 2020-10-31 | 0 | 2023-07-12 |
This allows us to make maps using the same functions used for other COVIDcast data:
plot(hospitalized, plot_type = "choro")Warning: Metadata for signal mean and standard deviation not available;
defaulting to observed mean and standard deviation to set plot range.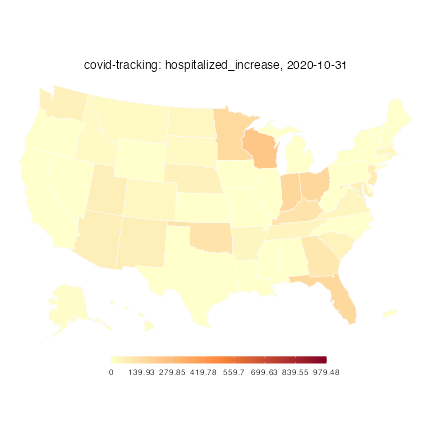
Analysis alongside other signals
Now that our data is loaded as a covidcast_signal
object, we can use it alongside other signals from the API. For example,
let’s examine how new COVID hospitalizations correlate with outpatient
doctor visits with deaths during October 2020, where we use death data
as reported by the API.
deaths <- covidcast_signal("jhu-csse", "deaths_7dav_incidence_prop",
start_day = "2020-10-01",
end_day = "2020-10-31",
geo_type = "state")
covidcast_cor(deaths, hospitalized, by = "time_value")# A tibble: 31 × 2
time_value value
<date> <dbl>
1 2020-10-01 0.218
2 2020-10-02 0.120
3 2020-10-03 0.147
4 2020-10-04 0.148
5 2020-10-05 0.0436
6 2020-10-06 0.0706
7 2020-10-07 0.123
8 2020-10-08 0.0300
9 2020-10-09 0.0955
10 2020-10-10 0.178
# ℹ 21 more rowsWe can also use the tools provided by this package to place both
signals into a single data frame for analysis. For example, to build a
model that uses hospitalizations and other data to predict deaths, it
may be convenient to produce a data frame where each row represents one
state on one day, and each column is a variable (such as hospitalization
or death). Using aggregate_signals(), this is easy:
death_hosp <- aggregate_signals(list(deaths, hospitalized),
format = "wide")
head(death_hosp) %>%
knitr::kable()| geo_value | time_value | value+0:jhu-csse_deaths_7dav_incidence_prop | value+0:covid-tracking_hospitalized_increase |
|---|---|---|---|
| ca | 2020-10-01 | 0.2286116 | 0 |
| ca | 2020-10-02 | 0.2137337 | 0 |
| ca | 2020-10-03 | 0.2122822 | 0 |
| ca | 2020-10-04 | 0.2122822 | 0 |
| ca | 2020-10-05 | 0.2177254 | 0 |
| ca | 2020-10-06 | 0.1788977 | 0 |